Publication Annotations
tools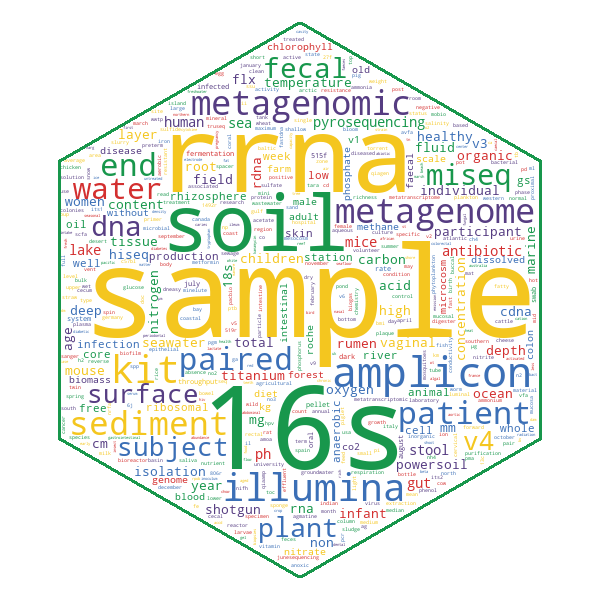 Automated annotations are now available for publications linked to metagenomic studies on MGnify, powered by Europe PMC.
Automated annotations are now available for publications linked to metagenomic studies on MGnify, powered by Europe PMC.
Annotations on MGnify Publications – powered by Europe PMC
Europe PMC’s annotations use text mining to search through biological and medical research publications, finding mentions of known entities like diseases, chemicals, protein interactions, locations, and environments. Multiple groups provide the text mining algorithms and annotations, with each provider having a different specialism.
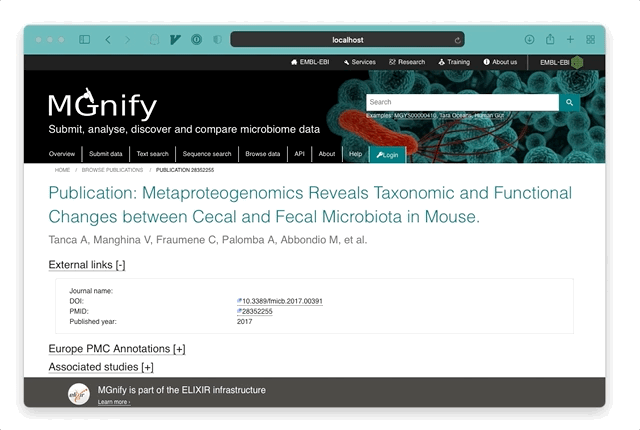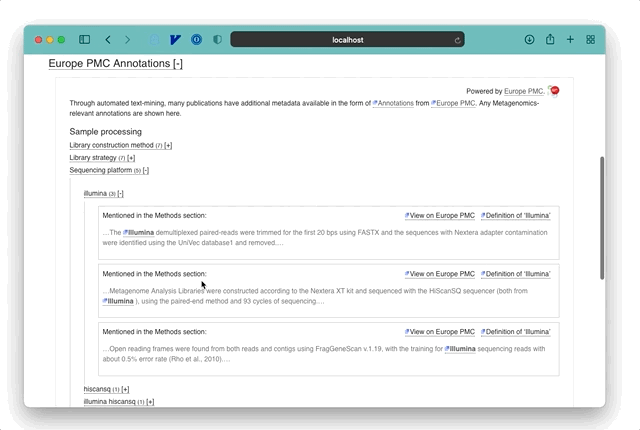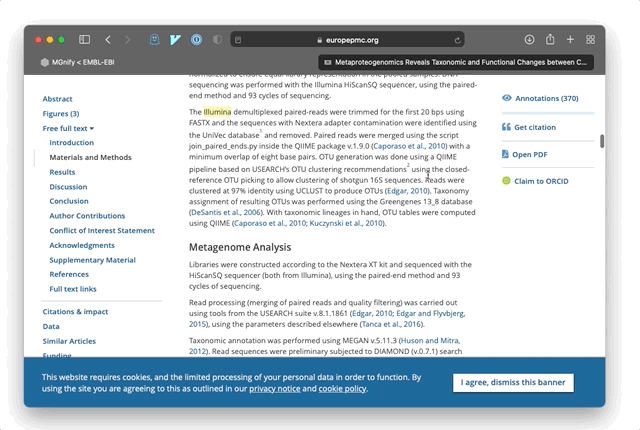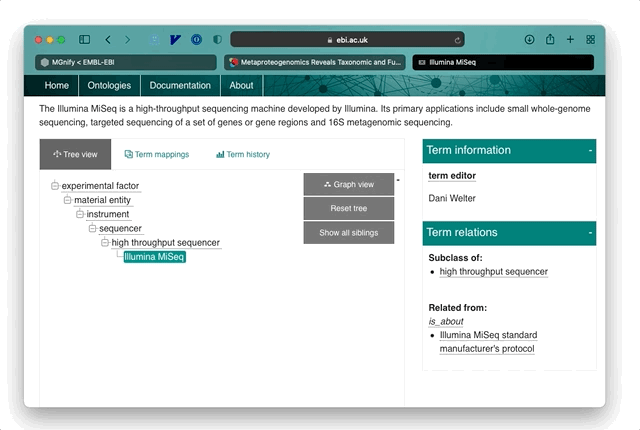
The EMERALD (Enriching MEtagenomics Results using Artificial intelligence and Literature Data) project is one of the 10 providers currently supported and focusses on annotating metagenomic terms. These cover the experimental methods such as the sequencing platform used, as well as other useful metadata like the Host environment a sample was taken from.

For example, if a paper mentions: “A sample was prepared for Illumina sequencing”, then EMERALD identifies the term ‘Illumina’ as a potential sequencing platform and stores an annotation pointing to the location of that phrase in the publication. Combined with open-access literature, these allow a new and rapid approach to exploring metagenomic data and research. The power of these annotations is in explicating a summary of the sample data and methods for a publication, beyond any formal keywords and metadata entered by authors into literature databases. In most cases, annotations are linked directly to the relevant section of a paper so that users can jump to the full text and understand the method or samples in more detail.
Within MGnify, these complement the snapshots of metadata derived from ENA and BioSamples provided on the sample pages.
The improved Publications pages of the MGnify website lists these Annotations alongside any related Samples.
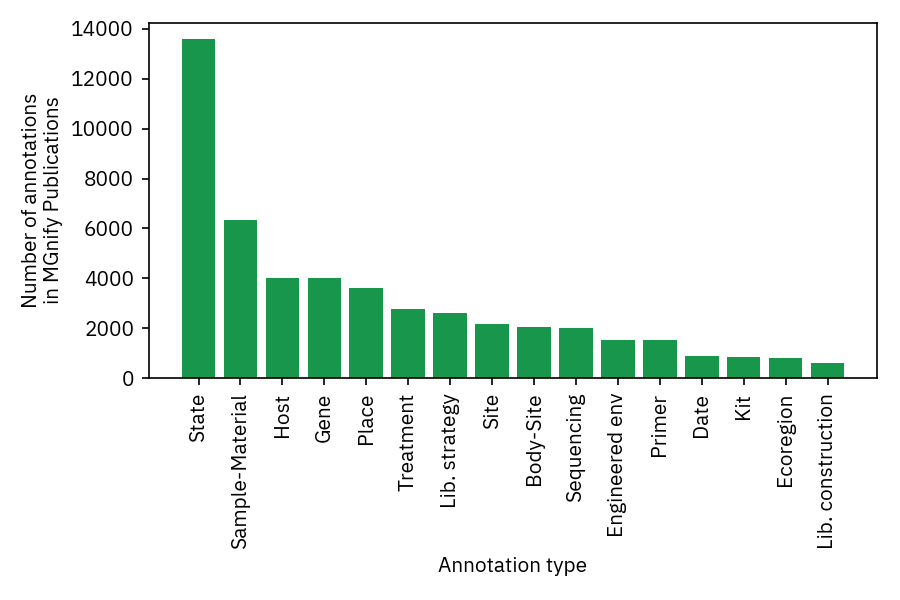
At the time of writing (November 2021), 625/869 of the publications linked in MGnify have metagenomic annotations available. An average publication with available annotations has 69 tags and paper extracts, which are visualised as a hierarchical tree in MGnify for easy browsing of the terms and their context.
The extracted terms cover a rich array of metagenomic metadata, and the most common annotation type is generally descriptions of samples.
Jump into this paper about Kombucha to try it out and let us know what you think.
