Anvi’o visualisations for refinement of Eukaryotic MAGs
 In the coming months we will be expanding MGnify Genomes to include Eukaryotic genomes. Take an early look at our plans to enable users to explore Eukaryotic genomes using Anvi’o.
In the coming months we will be expanding MGnify Genomes to include Eukaryotic genomes. Take an early look at our plans to enable users to explore Eukaryotic genomes using Anvi’o.

Articles
 In the coming months we will be expanding MGnify Genomes to include Eukaryotic genomes. Take an early look at our plans to enable users to explore Eukaryotic genomes using Anvi’o.
In the coming months we will be expanding MGnify Genomes to include Eukaryotic genomes. Take an early look at our plans to enable users to explore Eukaryotic genomes using Anvi’o.
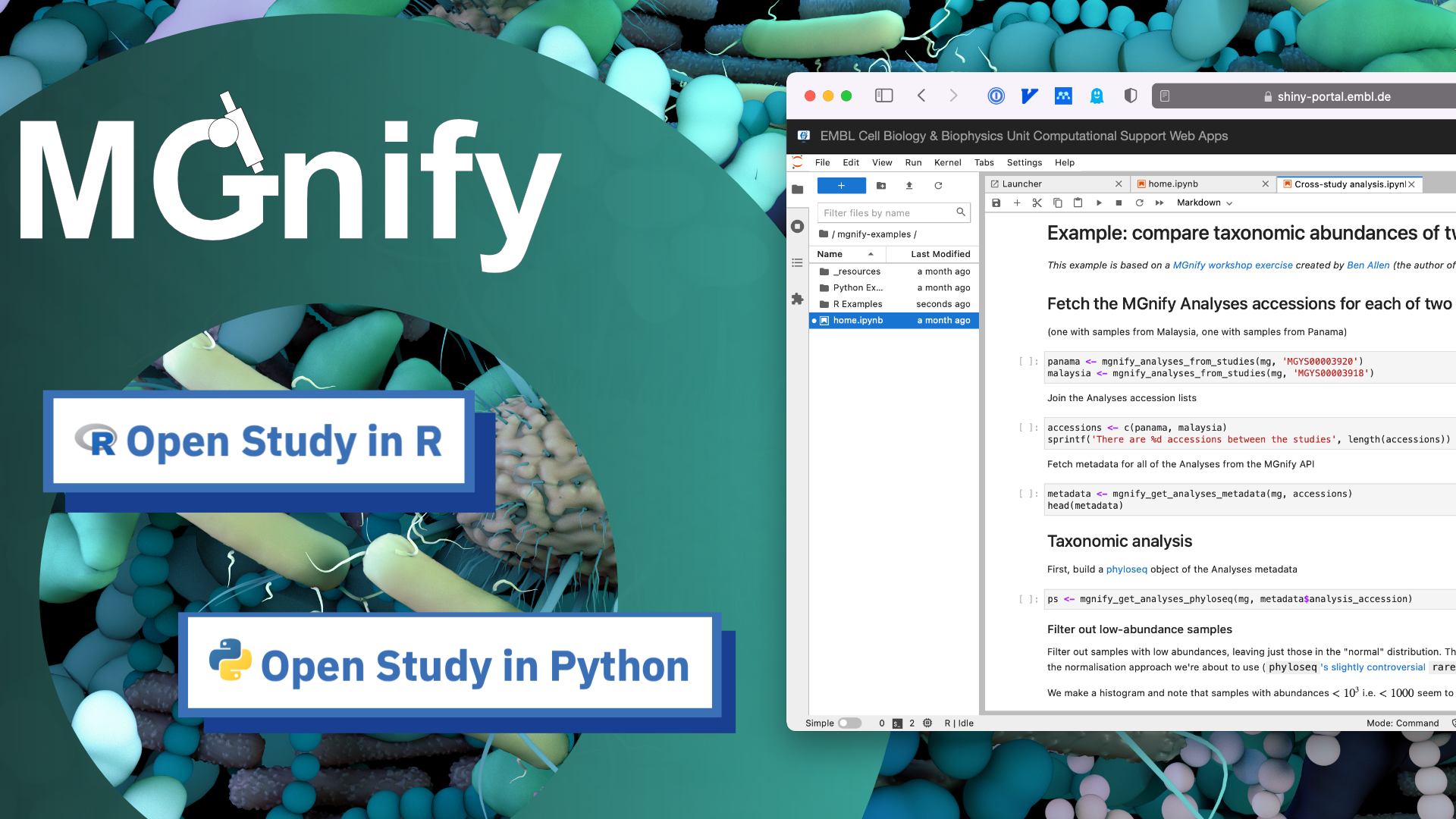 We are excited to announce the launch of MGnify’s Notebook Server. It provides an online, no-installation-needed, Jupyter Lab environment for users to explore programmatic access to MGnify’s datasets using Python or with R using the MGnifyR package.
We are excited to announce the launch of MGnify’s Notebook Server. It provides an online, no-installation-needed, Jupyter Lab environment for users to explore programmatic access to MGnify’s datasets using Python or with R using the MGnifyR package.
 Automated annotations are now available for publications linked to metagenomic studies on MGnify, powered by Europe PMC.
Automated annotations are now available for publications linked to metagenomic studies on MGnify, powered by Europe PMC.
![]() The MGnify protein sequence database comprises sequences predicted from assemblies generated from publicly available metagenomic datasets. The initial release in August 2017 comprised just under 50 million sequences; the current version contains in excess of 800 million. All sequences now have stable accessions.
The MGnify protein sequence database comprises sequences predicted from assemblies generated from publicly available metagenomic datasets. The initial release in August 2017 comprised just under 50 million sequences; the current version contains in excess of 800 million. All sequences now have stable accessions.
 Need to compile metadata to perform trait associations using our metagenomic data? Interested in correlating species abundance with the origin of the sample to identify organisms associated with a particular environment or state? Try our latest metagenomics toolkit (called: “mg-toolkit”) - a beta version of a tool to enable scientists to download all of the sample metadata for a given study to a single csv file. Simply install as follows:
Need to compile metadata to perform trait associations using our metagenomic data? Interested in correlating species abundance with the origin of the sample to identify organisms associated with a particular environment or state? Try our latest metagenomics toolkit (called: “mg-toolkit”) - a beta version of a tool to enable scientists to download all of the sample metadata for a given study to a single csv file. Simply install as follows:
 Want to perform comprehensive meta-analysis of samples from publicly available metagenomics studies? Interested in discovering patterns in metagenomic data to predict disease? Our REST API allows both human and machines to query over 100,000 publicly available metagenomic and metatranscriptomic datasets. The base URL to the API https://www.ebi.ac.uk/metagenomics/api/ provides access to several data collections, such as studies, samples, runs, biomes and experiment-types. They can be filtered by a set of attributes, such as biome, allowing selection of samples that belong to the same microbial ecosystem. For instance, to retrieve oceanic data: https://www.ebi.ac.uk/metagenomics/api/latest/studies?lineage=root:Environmental:Aquatic:Marine:Oceanic. Retrieving data from our API is as simple as sending an HTTP request, where the response returns a JSON object formatted data structure that contains the resource type, associated object identifier (id) with attributes and relationships linking to other resources. For example, https://www.ebi.ac.uk/metagenomics/api/latest/studies/PRJEB1787 retrieves a metagenomics dataset produced during experiments of the Tara Oceans Expedition.
Want to perform comprehensive meta-analysis of samples from publicly available metagenomics studies? Interested in discovering patterns in metagenomic data to predict disease? Our REST API allows both human and machines to query over 100,000 publicly available metagenomic and metatranscriptomic datasets. The base URL to the API https://www.ebi.ac.uk/metagenomics/api/ provides access to several data collections, such as studies, samples, runs, biomes and experiment-types. They can be filtered by a set of attributes, such as biome, allowing selection of samples that belong to the same microbial ecosystem. For instance, to retrieve oceanic data: https://www.ebi.ac.uk/metagenomics/api/latest/studies?lineage=root:Environmental:Aquatic:Marine:Oceanic. Retrieving data from our API is as simple as sending an HTTP request, where the response returns a JSON object formatted data structure that contains the resource type, associated object identifier (id) with attributes and relationships linking to other resources. For example, https://www.ebi.ac.uk/metagenomics/api/latest/studies/PRJEB1787 retrieves a metagenomics dataset produced during experiments of the Tara Oceans Expedition.
 Interested in bulk download of our data? Did you know that we provide a Python script for the bulk download of publicly available project data? The tool iterates over all samples and runs in a project and builds an appropriate root URL, which it uses to download individual analyses result files. Different file types can be specified, allowing you to download, for example, all reads encoding 16S rRNAs, all taxonomic assignments, or all predicted protein coding sequences, for a particular project. To find out more, click the ‘Bulk download script’ link below.
Interested in bulk download of our data? Did you know that we provide a Python script for the bulk download of publicly available project data? The tool iterates over all samples and runs in a project and builds an appropriate root URL, which it uses to download individual analyses result files. Different file types can be specified, allowing you to download, for example, all reads encoding 16S rRNAs, all taxonomic assignments, or all predicted protein coding sequences, for a particular project. To find out more, click the ‘Bulk download script’ link below.
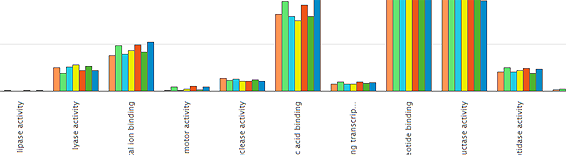 Interested in comparing the functional profile of sequencing runs within a project? Now it is possible, using our comparison tool, which provides analysis based on a slimmed-down subset of Gene Ontology (GO) terms, specially developed to describe metagenomic data.
Interested in comparing the functional profile of sequencing runs within a project? Now it is possible, using our comparison tool, which provides analysis based on a slimmed-down subset of Gene Ontology (GO) terms, specially developed to describe metagenomic data.