MGnify’s Notebook Server with MGnifyR launched
tools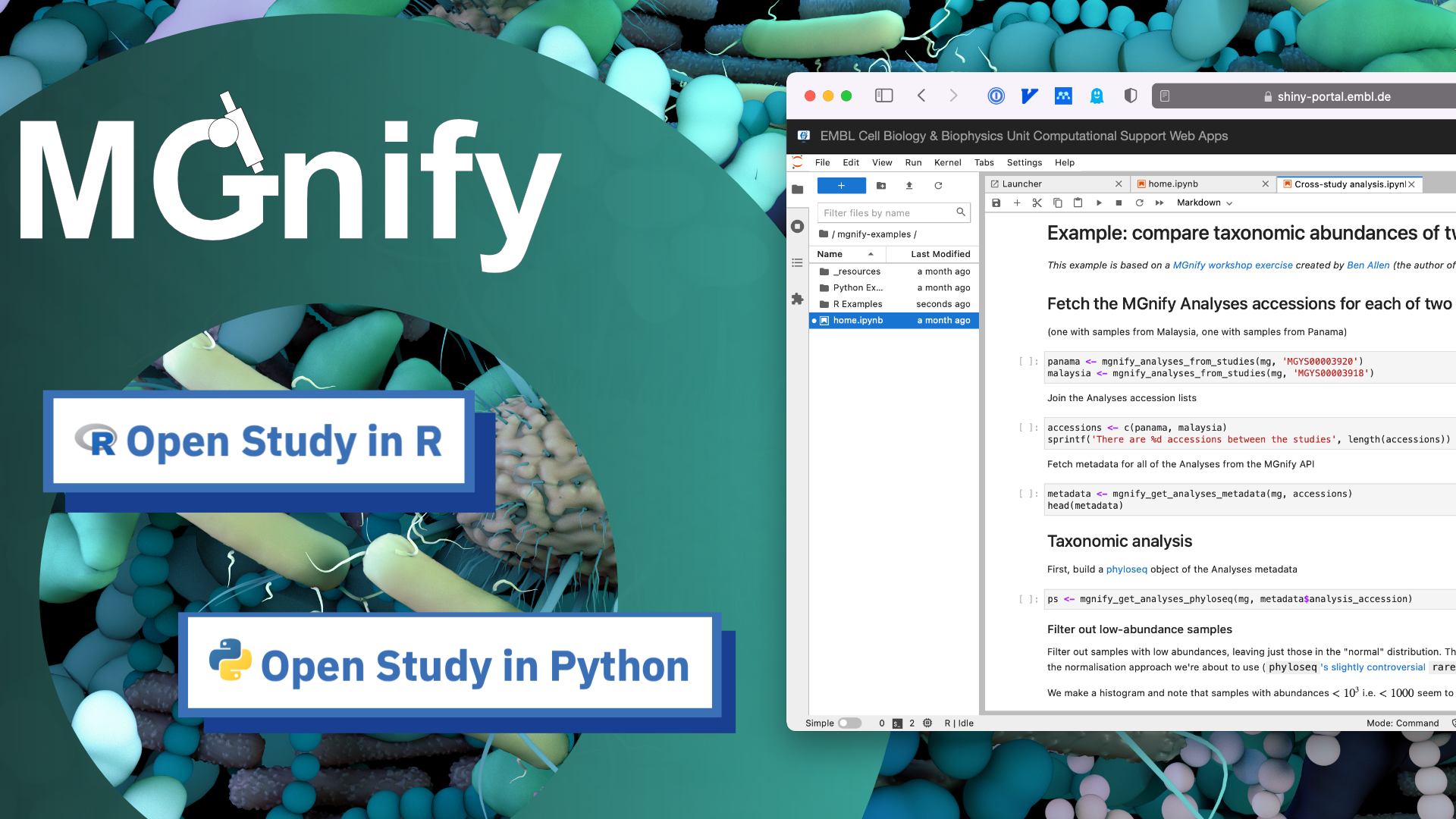 We are excited to announce the launch of MGnify’s Notebook Server. It provides an online, no-installation-needed, Jupyter Lab environment for users to explore programmatic access to MGnify’s datasets using Python or with R using the MGnifyR package.
We are excited to announce the launch of MGnify’s Notebook Server. It provides an online, no-installation-needed, Jupyter Lab environment for users to explore programmatic access to MGnify’s datasets using Python or with R using the MGnifyR package.
Watch our video tutorial on YouTube.
The quantity and richness of metagenomics-derived data in MGnify grows every day. The MGnify website is the best place to start exploring and searching the MGnify database, and allows users to download modest query results as CSV tables.
For larger queries, or more complex requirements like fetching metadata from samples across multiple studies, a programmatic access approach is far better.
 MGnify database entries for selected biomes
MGnify database entries for selected biomes
Programmatic access – fetching data from MGnify using a terminal command or code script – uses the MGnify API (Application Programming Interface). The API provides access to every data type in MGnify: Studies, Samples, Analyses, Annotations, MAGs etc: it is what lies behind the MGnify website. Using the API means you can fetch more data than is possible via the website, and can help you write reproducible analysis scripts.
The API can be explored interactively online, using the API Browser. But actually using the API first requires knowledge and/or installation of tools on your computer. This might range from a command line tool like cURL, to learning R and setting up the R Studio application, to setting up a Python environment and installing a suite of packages used for data analysis. Second, the API returns most data in JSON format: this is standard on the web, but less familiar for bioinformaticians used to TSVs and dataframes.
The MGnify Notebook Server and MGnifyR package are designed to bridge these gaps. Users can launch an online R and Python coding environment in their browser, without installing anything. The environment is hosted by EMBL’s Cell Biology and Biophysics Computational Support team, who support computational projects across EMBL. It already includes the main libraries needed for communicating with the MGnify API, analysing data, and making plots. It uses the popular Jupyter Lab software, which means you can code inside Notebooks: interactive code documents.
There are example Notebooks written in both R and Python, so users can pick whichever they’re more familiar with.
For many users, the online environment can serve as an extension of the website: enabling them to call a few queries on the API, join some data together, and create a datafile or plot they can save to their own computer. The MGnify website has deep-links into the Notebook Server that let users jump from looking at a Study on the website, for example, into a pre-configured R Notebook ready to access that Study using the API.
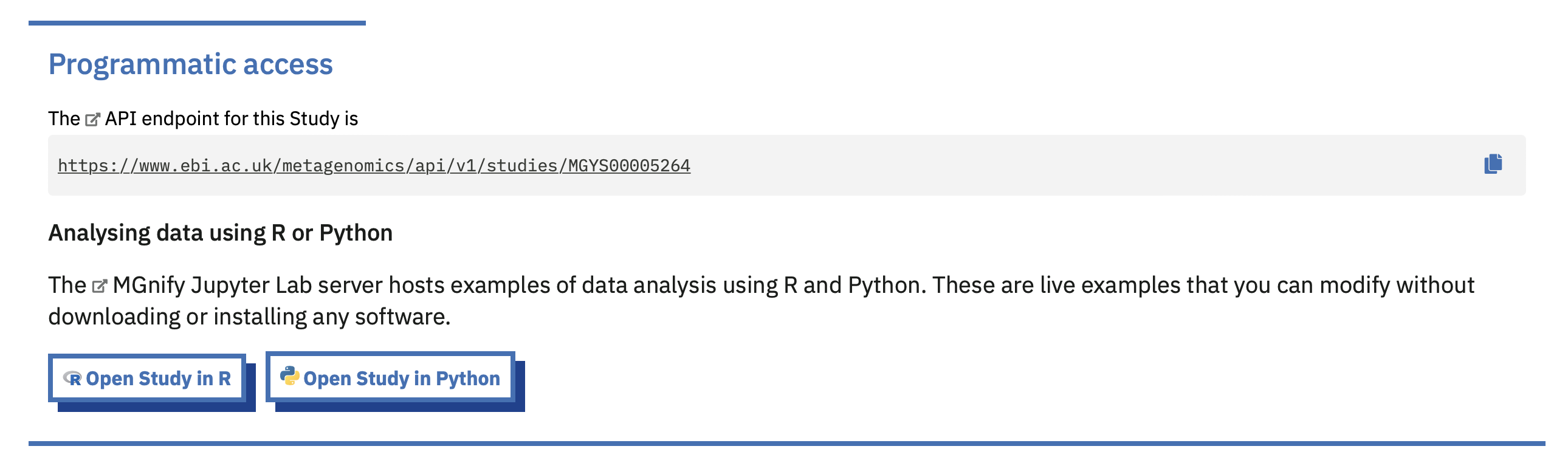 (This feature is available on the MGnify Beta website.)
(This feature is available on the MGnify Beta website.)
For more advanced use cases, the Notebooks serve as an introduction to using the API, and guide users on how to set up a local environment to use the API.
MGnifyR
The MGnify Notebook Server also provides access and examples for the MGnifyR package, created by Ben Allen. MGnifyR wraps the MGnify API in R functions that will feel more familiar to R users and translates data from the API’s JSON standard format into dataframes and Phyloseq objects. It also provides features for cross-study analysis workflows, like computing differential abundance.
