Cow rumen v1.0 MAG catalogue released
spotlight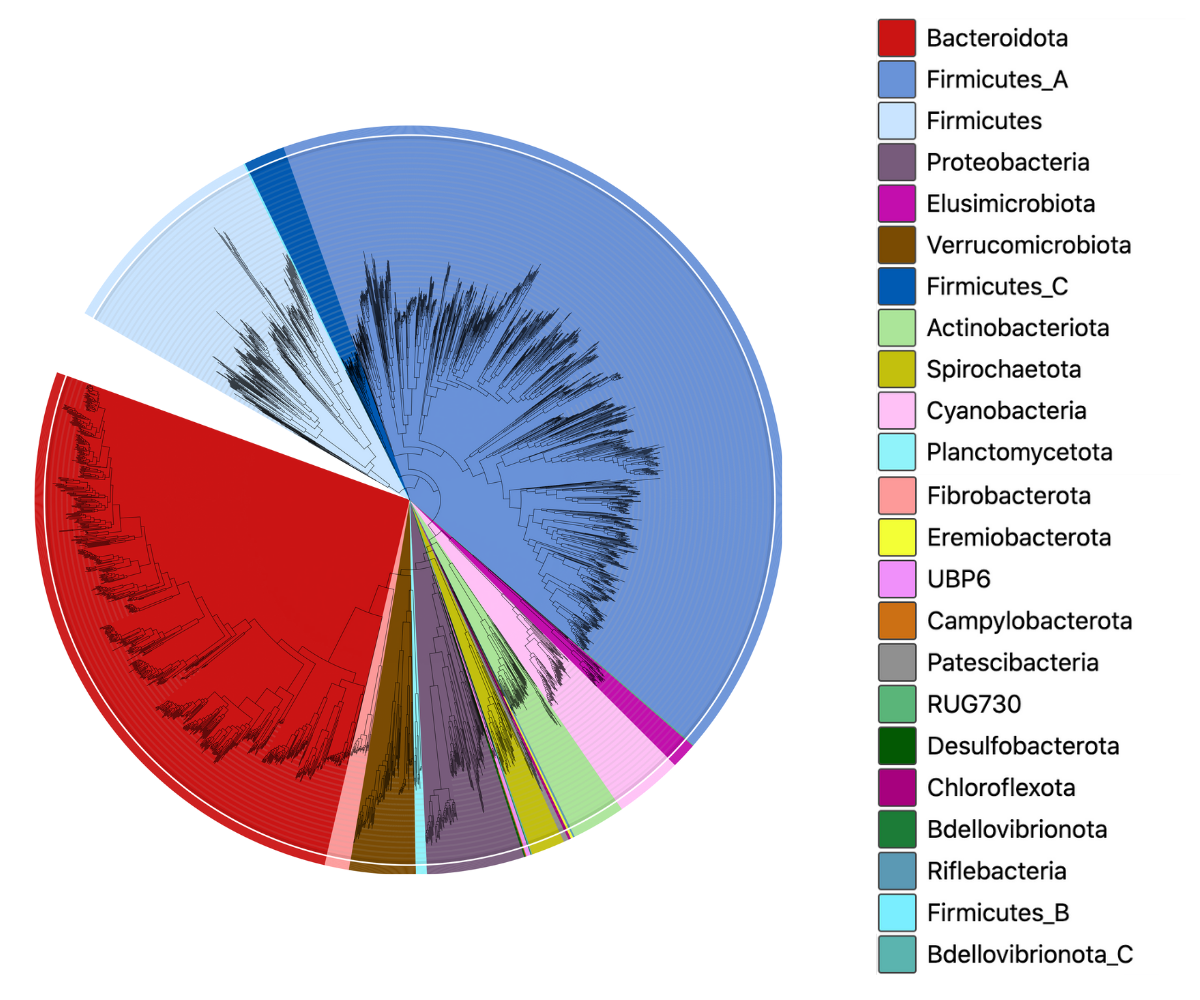 MGnify is delighted to announce the release of our latest MAG catalogue, comprising 2729 species-level cluster representative genomes derived from cow rumen datasets.
MGnify is delighted to announce the release of our latest MAG catalogue, comprising 2729 species-level cluster representative genomes derived from cow rumen datasets.
This catalogue is based on the Watson-lab rumen-uncultured genomes (RUGs) set published by Stewart et al. Nature Biotech (2019), and was generated as part of a collaboration between MGnify and the Watson-lab funded by BBSRC. The catalogue contains 5588 genomes from both the RUGs set as well as a large African cattle rumen-specific MAG set. To ensure compatibility across our MAG catalogues, the genomes were quality-filtered, dereplicated and clustered using the same parameters as our existing catalogues. The resulting taxonomic diversity can be seen in the image here:

As with our existing MAG catalogues, we have generated an associated protein catalogue which is available from our FTP site clustered at 100%, 95%, 90% and 50% amino acid identity.
We also provide search tools to compare your own data against this new catalogue. To compare a gene sequence against the catalogue we provide a BIGSI-based search, and to compare a whole genome or set of genomes you can use the Sourmash-based search.
We are particularly thrilled to be presenting the first MGnify MAG catalogue to incorporate publically available third-party generated MAGs, as this represents a scalable future for MAG identification. Availability of data within public repositories is a crucial aspect of open science, and this dataset is only available as a resource thanks to those researchers out there who submit their data to public resources such as ENA, and also to those bioinformatics researchers who subsequently submit their assemblies and MAGs to public repositories. We thank them all! Please let us know if you have any feedback on this new catalogue or any other aspect of MGnify using our helpdesk.
