Human gut v2.0
spotlight
Version 2 of the Unified Human Gastrointestinal Genome catalogue released
MGnify are excited to announce the release of version 2 of the Unified Human Gastrointestinal Genome (UHGG) catalogue. This is an updated version of the catalogue published by Almeida et al. Nature Biotech (2021). We have added 5,878 new genomes from two studies (PRJEB37358 and PRJNA544527), representing 129 new species.
Notably, this includes the set of isolate genomes from the most recent human gut culture collection generated by Poyet et al. Nature Medicine (2019). This means that the catalogue now contains a total of 289,232 prokaryotic genomes from the human gut microbiome clustered into 4,744 species representatives.
With the new genome additions, we have replaced 132 species representatives with better quality genomes (either a better quality MAG (metagenome assembled genome) or an isolate genome). To further increase the quality of the catalogue we have implemented the use of GUNC (a tool developed by the Bork group at EMBL) for genome quality filtering. Any singleton species (i.e., represented by only one strain) <90% complete flagged by GUNC as chimeric was excluded from the catalogue. Further details on the species replaced or excluded based on these criteria can be found in the associated README. We have also updated our genome accessions to use a standardised MGYG prefix, as well as a .version suffix to indicate an update to the genome sequence (such as for the removal of host contamination contigs).
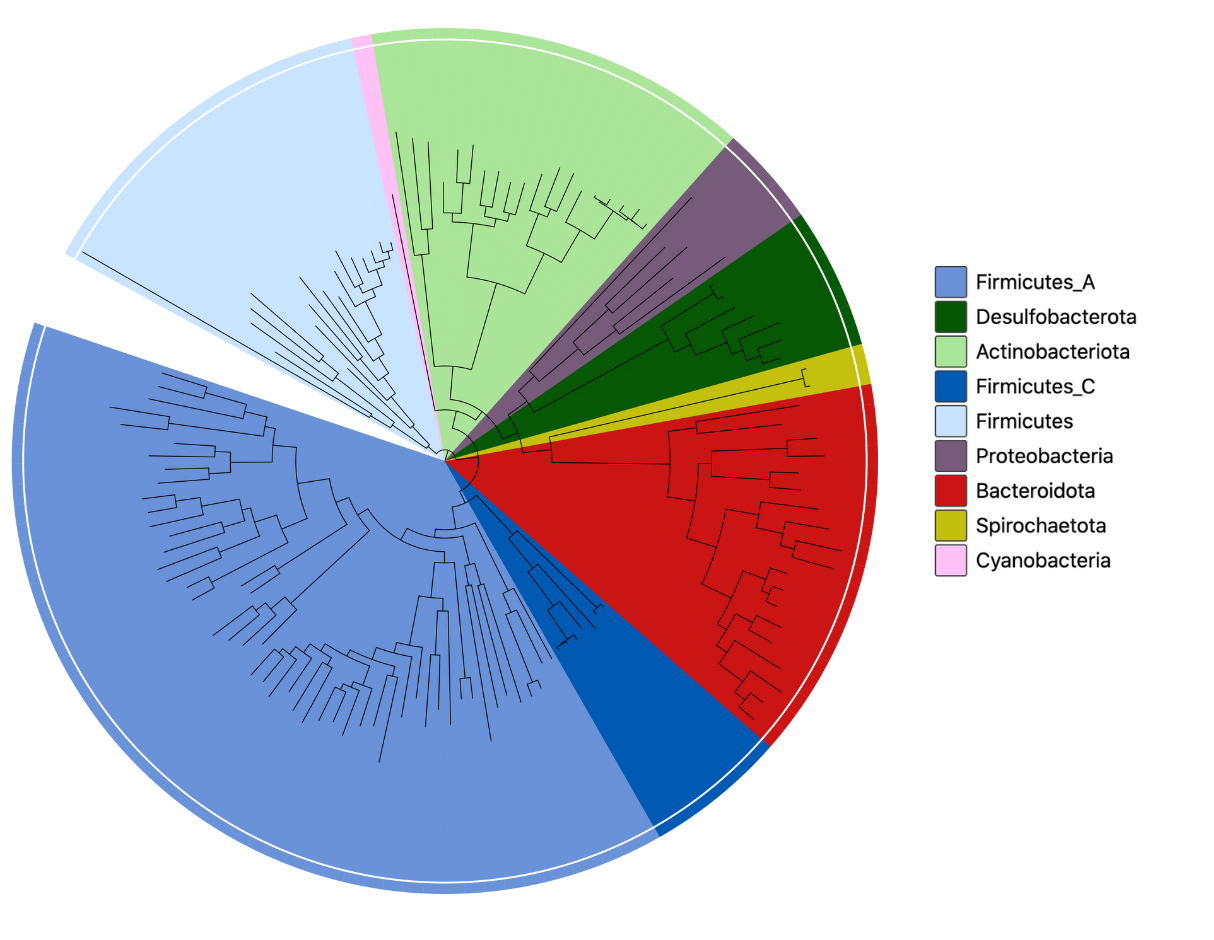
As for v1.0 we have also generated pan-genomes and an associated protein catalogue from all the genomes. However, we have replaced the use of Roary with Panaroo, due to its increased stringency for generating pan-genomes from MAGs. Protein-coding sequences were annotated with updated versions of eggNOG and InterPro.
For those who are relying on v1.0 for their ongoing projects do not worry: all the data from the former version is still available on our FTP, and a mapping between the accession formats is provided in genomes-all_metadata.tsv.
Moving forward, we will continue to provide regular updates to our existing catalogues based on the availability and number of new genomes released, and technical advancements in the field. We hope you find these data useful and feel free to provide us with any feedback you have.
