MGnify Genomes mouse gut catalogue v1.0 released
spotlight![]() We are thrilled to be presenting the latest MGnify Genomes catalogue comprising 112,951 genomes derived from mouse gut datasets, represented by 2,847 species-level cluster representative genomes.
We are thrilled to be presenting the latest MGnify Genomes catalogue comprising 112,951 genomes derived from mouse gut datasets, represented by 2,847 species-level cluster representative genomes.
This catalogue was generated as part of our work with the MRC funded National Mouse Genetics Network (NMGN), and combines three existing mouse gut genome catalogues, as well as genomes generated by MGnify from additional publicly-available datasets. The contributing catalogues are the Mouse Gastrointestinal Bacteria Catalogue (MGBC), the Comprehensive Mouse Microbiota Genome (CMMG) catalogue, and the integrated Mouse Gut Metagenomic Catalog (iMGMC) as published in Beresford-Jones et al. Cell Host & Microbe (2022), Kieser et al. PLoS Comput Biol (2022), and Lesker et al. Cell Rep (2020) respectively.
The total genomes in the catalogue were quality-filtered and clustered using the catalogue generation process as described in Gurbich et al. (2023). An overview of the process is shown in Figure 1 and the resulting taxonomic diversity of the mouse gut catalogue is represented in Figure 2.
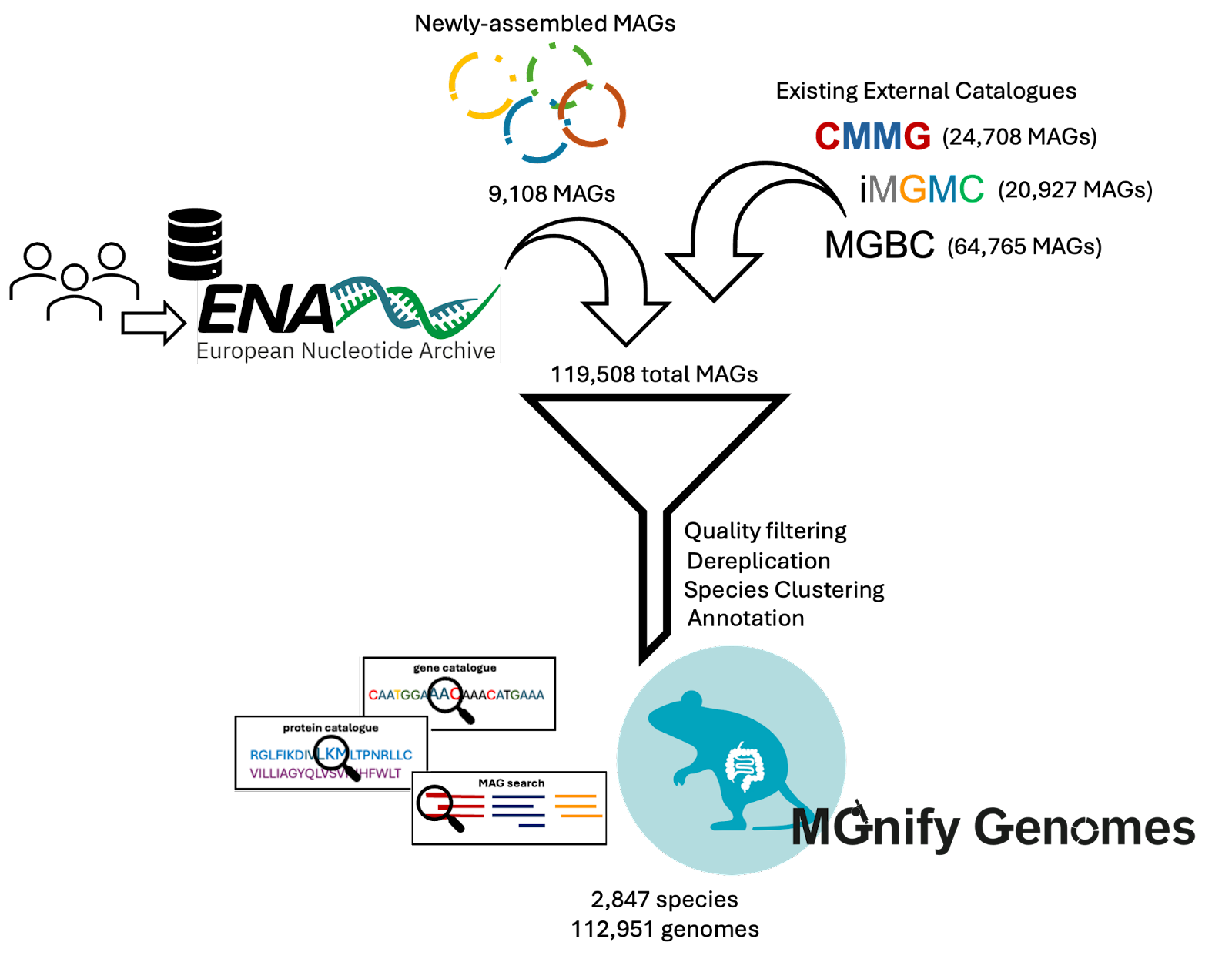
Figure 1: Overview of catalogue generation. The mouse gut catalogue was generated from a combination of newly-assembled MAGs derived from publicly-available Whole Genome Shotgun (WGS) datasets, and existing genomes from three external catalogues. The MGnify Genomes pipeline (as described in Gurbich et al. (2023)) was used to produce the final mouse gut catalogue.
As with existing MGnify Genome catalogues, we provide an associated protein catalogue, available from our FTP site, clustered at 100%, 95%, 90% and 50% amino acid identity. MGnify Genomes’ search tools enable users to compare their own data against the catalogue. Gene sequences can be searched against the catalogue using a COBS-based search, whereas whole genomes or sets of genomes can be compared using a sourmash-based search.
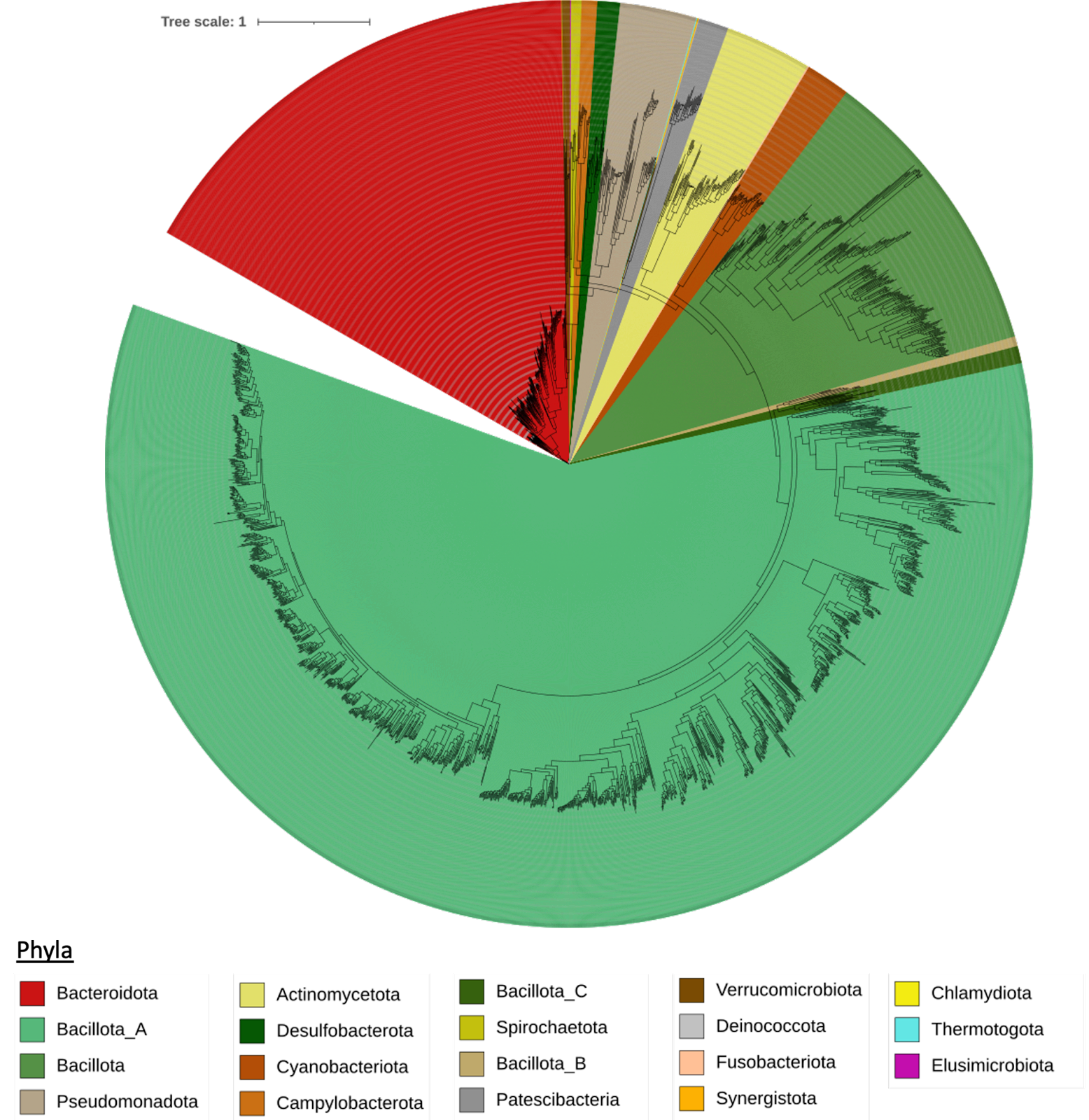
Figure 2: Taxonomic diversity of mouse gut catalogue. A phylogenetic tree showing the diversity of the genomes in the mouse gut catalogue with taxonomy assigned using GTDB r214, and coloured at the Phyla level.
Future focus
Within the NMGN, some of the use-cases of this catalogue are to measure the variability in gut microbiomes between different mouse facilities, and to identify taxonomic and functional features that are associated with disease states or conditions of interest. More broadly this catalogue will support translational research between mouse and human: to what extent can the mouse and human microbiota be compared in terms of functional and taxonomic summaries.
Generating this catalogue relied on the availability of data within public repositories, and so was only possible thanks to the many researchers who have submitted their data to public resources such as the European Nucleotide Archive (ENA). We gratefully acknowledge everyone who has submitted their data to these resources, and also the MRC National Mouse Genetics Network for their generous funding. Any feedback on the new catalogue or any other aspect of the MGnify resource would be greatly appreciated and can be submitted through our helpdesk.
